61 changed files with 350 additions and 0 deletions
BIN
code/fonctions_activations_classiques_bis/images_lstm/0.0015245312824845314_1_25_0.png
View File
BIN
code/fonctions_activations_classiques_bis/images_lstm/0.0032665664330124855_2_23_0.png
View File
BIN
code/fonctions_activations_classiques_bis/images_lstm/0.007591465953737497_0_22_2.png
View File
BIN
code/fonctions_activations_classiques_bis/images_lstm_complexe/0.0033504192251712084_1_20_0.png
View File
BIN
code/fonctions_activations_classiques_bis/images_lstm_complexe/0.006223898380994797_3_24_1.png
View File
BIN
code/fonctions_activations_classiques_bis/images_lstm_complexe/0.10564947128295898_0_23_0.png
View File
BIN
code/fonctions_activations_classiques_bis/images_lstm_complexe/0.12378907203674316_2_32_0.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.06567463278770447_18_68_1_0.22671216968856572.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.07205970585346222_21_124_1_0.2367283953038779.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.08117233216762543_17_65_1_0.21888422513950936.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.09494179487228394_16_126_1_0.2185005159425904.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.11478365957736969_6_257_0_0.045970875282274726.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.11892988532781601_15_175_0_0.21927221765888907.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.12665440142154694_11_303_0_0.005309218769096553.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.1270594596862793_2_503_0_0.0018359787728027376.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.12761712074279785_13_222_0_0.04459920400317374.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.12833552062511444_4_204_0_0.29203033007606816.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.12948699295520782_22_69_1_0.25079012385640953.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.13041436672210693_1_437_0_0.14603480055924464.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.131441131234169_12_303_0_0.0768451657080157.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.13175156712532043_19_82_1_0.23020846993946503.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.1339569389820099_7_400_0_0.15216949883417233.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.18354983627796173_3_477_2_0.27801307355957433.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.18860067427158356_5_439_2_0.14623561250425976.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.19291751086711884_8_328_1_0.2827477811008291.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.19725225865840912_14_344_1_0.10697962339113726.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.1989164501428604_24_161_1_0.25351196870033027.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.19935593008995056_10_223_1_0.09378187159128668.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.20013734698295593_0_470_1_0.0164413606601999.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.2023385614156723_23_122_2_0.1977578401481706.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.20569606125354767_9_96_2_0.05150398613791677.png
View File
BIN
code/fonctions_activations_classiques_bis/images_relu/0.21392779052257538_20_144_1_0.18328780888516766.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.06598746031522751_2_320_2_0.1822093246051402.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.06716939061880112_3_408_1_0.05435687802020495.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.06960835307836533_11_459_1_0.15078379493470068.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07039721310138702_13_419_1_0.17673630086875358.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07124108821153641_12_254_1_0.08798038532486387.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07140277326107025_23_449_1_0.15069732052561416.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07182151824235916_18_506_1_0.12393136139749764.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07227852195501328_10_268_1_0.29715677189910183.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07250253111124039_21_462_1_0.1463326335395087.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07254311442375183_22_456_1_0.1808634658836852.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07295650988817215_15_340_1_0.1867230805414493.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07320355623960495_20_380_1_0.21169586538635998.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07320578396320343_0_314_1_0.2524618193850152.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07744359225034714_1_361_0_0.22495553592208314.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07764129340648651_7_410_0_0.010014353235764228.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07782604545354843_24_433_0_0.1249179581286838.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07789068669080734_17_232_0_0.05035593093863518.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.07806289196014404_6_137_0_0.152499022020141.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.11341426521539688_16_403_2_0.11854245911158873.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.13494135439395905_19_304_2_0.04651067517409238.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.13946005702018738_9_352_2_0.09476580480062193.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.1475277841091156_14_204_2_0.05430878580389549.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.18465152382850647_4_496_2_0.01462186922817893.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.19307482242584229_8_69_2_0.2199772091965577.png
View File
BIN
code/fonctions_activations_classiques_bis/images_snake/0.20571038126945496_5_147_2_0.0870870477658672.png
View File
+ 79
- 0
code/fonctions_activations_classiques_bis/trian_sin.py
View File
| @@ -0,0 +1,79 @@ | |||
| import tensorflow as tf | |||
| import tensorflow_addons as tfa | |||
| from matplotlib import pyplot as plt | |||
| import numpy as np | |||
| import optuna | |||
| densite = 10 | |||
| start1 = 5 | |||
| stop1 = 15 | |||
| start2 = 35 | |||
| stop2 = 45 | |||
| start = -50 | |||
| stop = 150 | |||
| training = [np.concatenate((np.linspace(start1, stop1, (stop1-start1)*densite), | |||
| np.linspace(start2, stop2, (stop2-start2)*densite)))] | |||
| training.append(np.sin(training[0])/3+0.5) | |||
| training[0] = training[0]/(stop-start) | |||
| validation = [np.linspace(start, stop, (stop-start)*densite)] | |||
| validation.append(np.sin(validation[0])/3+0.5) | |||
| validation[0] = validation[0]/(stop-start) | |||
| fig = plt.figure(1) | |||
| ax1 = fig.add_subplot(2, 1, 1) | |||
| ax2 = fig.add_subplot(2, 1, 2, sharex=ax1) | |||
| ax1.plot(*validation,'.', label="validation") | |||
| ax2.plot(*training,'.', label="apprentissage") | |||
| plt.legend() | |||
| plt.show() | |||
| bce = tf.keras.losses.BinaryCrossentropy() | |||
| mse = tf.keras.losses.MeanSquaredError() | |||
| def objectif(trial) : | |||
| HIDDEN = trial.suggest_int('hidden',64,512) | |||
| SIZE = trial.suggest_int('size',0,2) | |||
| DROPOUT = trial.suggest_float('dropout', 0,0.3) | |||
| model = tf.keras.Sequential() | |||
| model.add(tf.keras.layers.Dense(HIDDEN, input_shape=(1,), activation=tfa.activations.snake)) | |||
| for i in range(SIZE) : | |||
| model.add(tf.keras.layers.Dense(HIDDEN, activation=tfa.activations.snake)) | |||
| model.add(tf.keras.layers.Dropout(DROPOUT)) | |||
| model.add(tf.keras.layers.Dense(1, activation='sigmoid')) | |||
| model.compile(optimizer='adam', | |||
| loss='bce', | |||
| metrics=['mse']) | |||
| history = model.fit(x=training[0], y=training[1], batch_size=4, | |||
| epochs=3000, shuffle=True, | |||
| #validation_data=(validation[0], validation[1]), | |||
| verbose='auto') | |||
| pred = model.predict(validation[0]) | |||
| plt.figure(2) | |||
| plt.clf() | |||
| plt.plot(*validation, label="validation") | |||
| plt.plot(*training, label="apprentissage") | |||
| plt.plot(validation[0], pred, label="prediction") | |||
| plt.legend() | |||
| rep = mse(pred-0.5,validation[1]-0.5).numpy() | |||
| plt.savefig(f"images_snake/{rep}_{trial.number}_{HIDDEN}_{SIZE}_{DROPOUT}.png") | |||
| return rep | |||
| study = optuna.create_study() | |||
| study.optimize(objectif, n_trials=25) | |||
+ 96
- 0
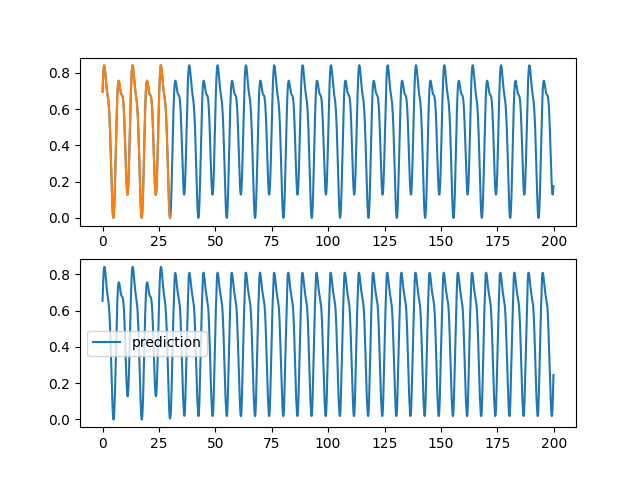
code/fonctions_activations_classiques_bis/trian_sin_lstm.py
View File
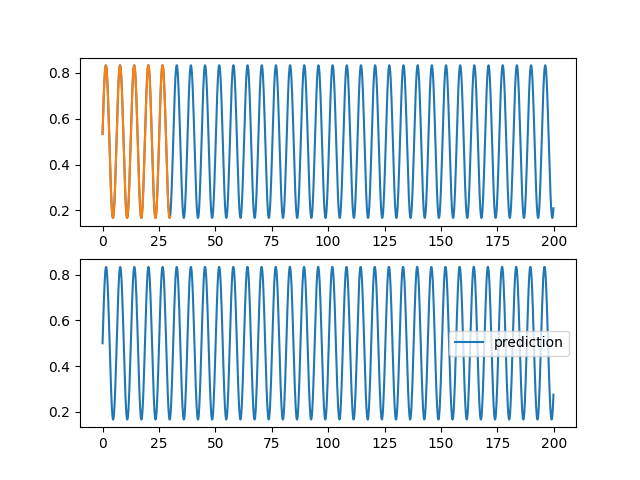
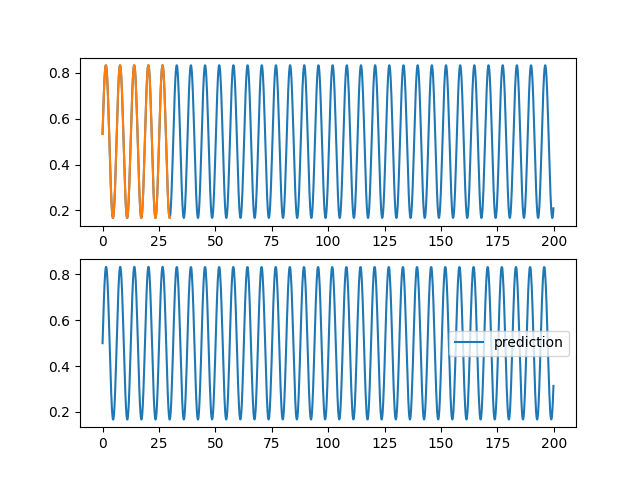
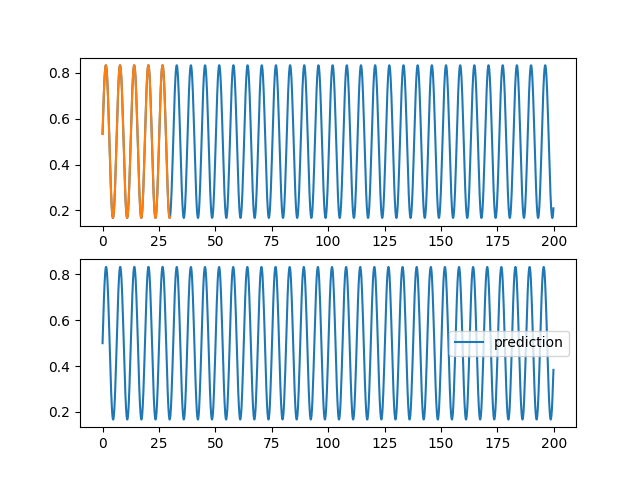
| @@ -0,0 +1,96 @@ | |||
| import tensorflow as tf | |||
| import tensorflow_addons as tfa | |||
| from matplotlib import pyplot as plt | |||
| import numpy as np | |||
| import optuna | |||
| densite = 10 | |||
| start1 = 0 | |||
| stop1 = 30 | |||
| start = 0 | |||
| stop = 200 | |||
| def f(x) : | |||
| return np.sin(x) | |||
| training = [np.linspace(start1, stop1, (stop1-start1)*densite)] | |||
| training.append(f(training[0])/3+0.5) | |||
| training = [training[0][:-1], training[1][:-1],training[1][1:]] | |||
| validation = [np.linspace(start, stop, (stop-start)*densite)] | |||
| validation.append(f(validation[0])/3+0.5) | |||
| validation = [validation[0][:-1], validation[1][:-1],validation[1][1:]] | |||
| fig = plt.figure(1) | |||
| ax1 = fig.add_subplot(2, 1, 1) | |||
| ax2 = fig.add_subplot(2, 1, 2, sharex=ax1) | |||
| ax1.plot(validation[0],validation[2], label="validation") | |||
| ax2.plot(training[0], training[2], label="apprentissage") | |||
| ax2.plot(training[0], training[1], label="apprentissage") | |||
| plt.legend() | |||
| plt.show() | |||
| training[1] = np.expand_dims(training[1], [0,2]) | |||
| training[2] = np.expand_dims(training[2], [0,2]) | |||
| validation[1] = np.expand_dims(validation[1], [0,2]) | |||
| validation[2] = np.expand_dims(validation[2], [0,2]) | |||
| bce = tf.keras.losses.BinaryCrossentropy() | |||
| mse = tf.keras.losses.MeanSquaredError() | |||
| def objectif(trial) : | |||
| HIDDEN = trial.suggest_int('hidden',16,32) | |||
| SIZE = trial.suggest_int('size',0,2) | |||
| model = tf.keras.Sequential() | |||
| model.add(tf.keras.layers.LSTM(HIDDEN, return_sequences=True)) | |||
| for i in range(SIZE) : | |||
| model.add(tf.keras.layers.LSTM(HIDDEN, return_sequences=True)) | |||
| model.add(tf.keras.layers.Dense(HIDDEN, activation='relu')) | |||
| model.add(tf.keras.layers.Dense(HIDDEN//2, activation='relu')) | |||
| model.add(tf.keras.layers.Dense(1, activation='sigmoid')) | |||
| model.compile(optimizer='adam', | |||
| loss='bce', | |||
| metrics=['mse']) | |||
| history = model.fit(x=training[1], y=training[2], | |||
| epochs=300, shuffle=True, | |||
| #validation_data=(validation[0], validation[1]), | |||
| verbose='auto') | |||
| pred = training[1] | |||
| while len(pred[0]) < len(validation[1][0]) : | |||
| print(len(validation[1][0]) - len(pred[0])) | |||
| pred = np.concatenate((pred,np.expand_dims(model.predict(pred)[0,-1],(1,2))), 1) | |||
| fig = plt.figure(1) | |||
| plt.clf() | |||
| ax1 = fig.add_subplot(2, 1, 1) | |||
| ax2 = fig.add_subplot(2, 1, 2, sharex=ax1) | |||
| ax1.plot(validation[0], validation[2][0,:,0], label="objectif") | |||
| ax1.plot(training[0], training[2][0,:,0], label="apprentissage") | |||
| plt.legend() | |||
| ax2.plot(validation[0], pred[0,:,0], label="prediction") | |||
| plt.legend() | |||
| rep = mse(pred,validation[2]).numpy() | |||
| plt.savefig(f"images_lstm/{rep}_{trial.number}_{HIDDEN}_{SIZE}.png") | |||
| return rep | |||
| study = optuna.create_study() | |||
| study.optimize(objectif, n_trials=25) | |||
+ 96
- 0
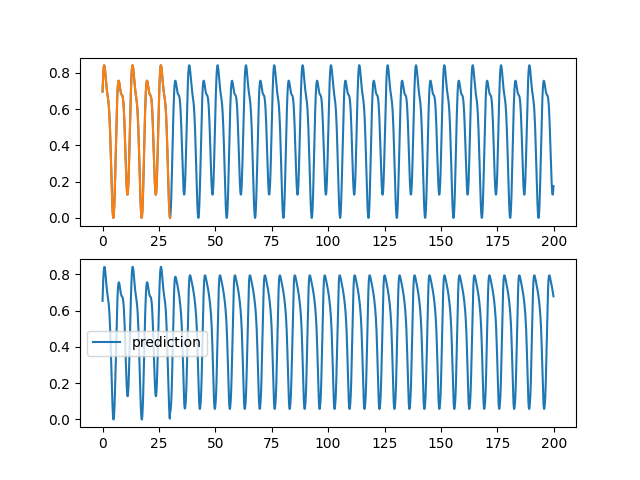
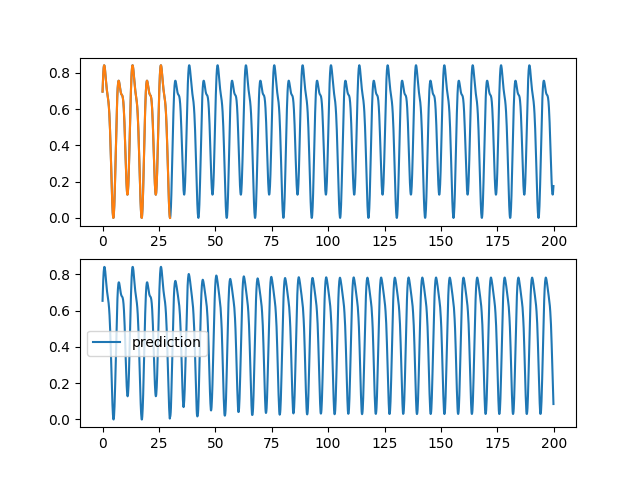
code/fonctions_activations_classiques_bis/trian_sin_lstm_complexe.py
View File
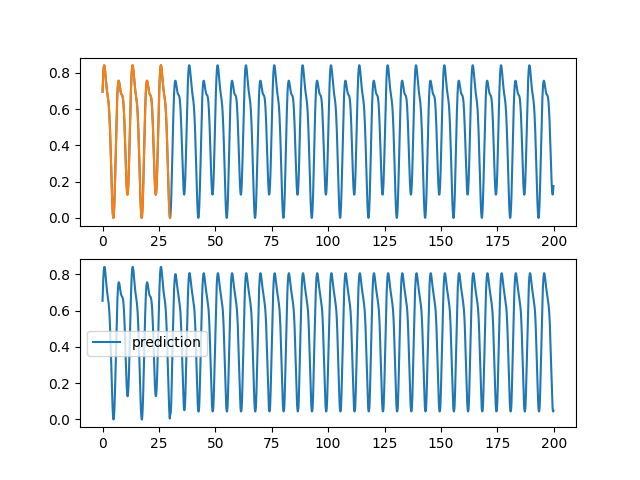
| @@ -0,0 +1,96 @@ | |||
| import tensorflow as tf | |||
| import tensorflow_addons as tfa | |||
| from matplotlib import pyplot as plt | |||
| import numpy as np | |||
| import optuna | |||
| densite = 10 | |||
| start1 = 0 | |||
| stop1 = 30 | |||
| start = 0 | |||
| stop = 200 | |||
| def f(x) : | |||
| return (np.sin(x) + 1/3*np.sin(2*x+1) + + 1/5*np.sin(x/2+2)) | |||
| training = [np.linspace(start1, stop1, (stop1-start1)*densite)] | |||
| training.append(f(training[0])/3+0.5) | |||
| training = [training[0][:-1], training[1][:-1],training[1][1:]] | |||
| validation = [np.linspace(start, stop, (stop-start)*densite)] | |||
| validation.append(f(validation[0])/3+0.5) | |||
| validation = [validation[0][:-1], validation[1][:-1],validation[1][1:]] | |||
| fig = plt.figure(1) | |||
| ax1 = fig.add_subplot(2, 1, 1) | |||
| ax2 = fig.add_subplot(2, 1, 2, sharex=ax1) | |||
| ax1.plot(validation[0],validation[2], label="validation") | |||
| ax2.plot(training[0], training[2], label="apprentissage") | |||
| ax2.plot(training[0], training[1], label="apprentissage") | |||
| plt.legend() | |||
| plt.show() | |||
| training[1] = np.expand_dims(training[1], [0,2]) | |||
| training[2] = np.expand_dims(training[2], [0,2]) | |||
| validation[1] = np.expand_dims(validation[1], [0,2]) | |||
| validation[2] = np.expand_dims(validation[2], [0,2]) | |||
| bce = tf.keras.losses.BinaryCrossentropy() | |||
| mse = tf.keras.losses.MeanSquaredError() | |||
| def objectif(trial) : | |||
| HIDDEN = trial.suggest_int('hidden',16,32) | |||
| SIZE = trial.suggest_int('size',0,2) | |||
| model = tf.keras.Sequential() | |||
| model.add(tf.keras.layers.LSTM(HIDDEN, return_sequences=True)) | |||
| for i in range(SIZE) : | |||
| model.add(tf.keras.layers.LSTM(HIDDEN, return_sequences=True)) | |||
| model.add(tf.keras.layers.Dense(HIDDEN, activation='relu')) | |||
| model.add(tf.keras.layers.Dense(HIDDEN//2, activation='relu')) | |||
| model.add(tf.keras.layers.Dense(1, activation='sigmoid')) | |||
| model.compile(optimizer='adam', | |||
| loss='bce', | |||
| metrics=['mse']) | |||
| history = model.fit(x=training[1], y=training[2], | |||
| epochs=300, shuffle=True, | |||
| #validation_data=(validation[0], validation[1]), | |||
| verbose='auto') | |||
| pred = training[1] | |||
| while len(pred[0]) < len(validation[1][0]) : | |||
| print(len(validation[1][0]) - len(pred[0])) | |||
| pred = np.concatenate((pred,np.expand_dims(model.predict(pred)[0,-1],(1,2))), 1) | |||
| fig = plt.figure(1) | |||
| plt.clf() | |||
| ax1 = fig.add_subplot(2, 1, 1) | |||
| ax2 = fig.add_subplot(2, 1, 2, sharex=ax1) | |||
| ax1.plot(validation[0], validation[2][0,:,0], label="objectif") | |||
| ax1.plot(training[0], training[2][0,:,0], label="apprentissage") | |||
| plt.legend() | |||
| ax2.plot(validation[0], pred[0,:,0], label="prediction") | |||
| plt.legend() | |||
| rep = mse(pred,validation[2]).numpy() | |||
| plt.savefig(f"images_lstm_complexe/{rep}_{trial.number}_{HIDDEN}_{SIZE}.png") | |||
| return rep | |||
| study = optuna.create_study() | |||
| study.optimize(objectif, n_trials=25) | |||
+ 79
- 0
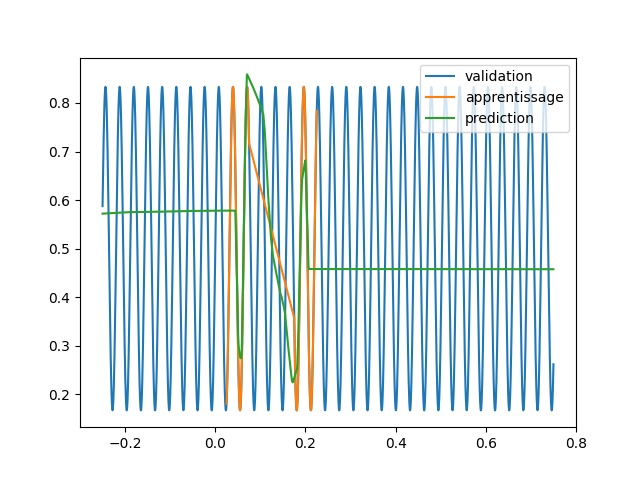
code/fonctions_activations_classiques_bis/trian_sin_relu.py
View File
| @@ -0,0 +1,79 @@ | |||
| import tensorflow as tf | |||
| import tensorflow_addons as tfa | |||
| from matplotlib import pyplot as plt | |||
| import numpy as np | |||
| import optuna | |||
| densite = 10 | |||
| start1 = 5 | |||
| stop1 = 15 | |||
| start2 = 35 | |||
| stop2 = 45 | |||
| start = -50 | |||
| stop = 150 | |||
| training = [np.concatenate((np.linspace(start1, stop1, (stop1-start1)*densite), | |||
| np.linspace(start2, stop2, (stop2-start2)*densite)))] | |||
| training.append(np.sin(training[0])/3+0.5) | |||
| training[0] = training[0]/(stop-start) | |||
| validation = [np.linspace(start, stop, (stop-start)*densite)] | |||
| validation.append(np.sin(validation[0])/3+0.5) | |||
| validation[0] = validation[0]/(stop-start) | |||
| fig = plt.figure(1) | |||
| ax1 = fig.add_subplot(2, 1, 1) | |||
| ax2 = fig.add_subplot(2, 1, 2, sharex=ax1) | |||
| ax1.plot(*validation,'.', label="validation") | |||
| ax2.plot(*training,'.', label="apprentissage") | |||
| plt.legend() | |||
| plt.show() | |||
| bce = tf.keras.losses.BinaryCrossentropy() | |||
| mse = tf.keras.losses.MeanSquaredError() | |||
| def objectif(trial) : | |||
| HIDDEN = trial.suggest_int('hidden',64,512) | |||
| SIZE = trial.suggest_int('size',0,2) | |||
| DROPOUT = trial.suggest_float('dropout', 0,0.3) | |||
| model = tf.keras.Sequential() | |||
| model.add(tf.keras.layers.Dense(HIDDEN, input_shape=(1,), activation='relu')) | |||
| for i in range(SIZE) : | |||
| model.add(tf.keras.layers.Dense(HIDDEN, activation='relu')) | |||
| model.add(tf.keras.layers.Dropout(DROPOUT)) | |||
| model.add(tf.keras.layers.Dense(1, activation='sigmoid')) | |||
| model.compile(optimizer='adam', | |||
| loss='bce', | |||
| metrics=['mse']) | |||
| history = model.fit(x=training[0], y=training[1], batch_size=4, | |||
| epochs=3000, shuffle=True, | |||
| #validation_data=(validation[0], validation[1]), | |||
| verbose='auto') | |||
| pred = model.predict(validation[0]) | |||
| plt.figure(2) | |||
| plt.clf() | |||
| plt.plot(*validation, label="validation") | |||
| plt.plot(*training, label="apprentissage") | |||
| plt.plot(validation[0], pred, label="prediction") | |||
| plt.legend() | |||
| rep = mse(pred-0.5,validation[1]-0.5).numpy() | |||
| plt.savefig(f"images_relu/{rep}_{trial.number}_{HIDDEN}_{SIZE}_{DROPOUT}.png") | |||
| return rep | |||
| study = optuna.create_study() | |||
| study.optimize(objectif, n_trials=25) | |||
Loading…
